Pathogenesis of Autism Spectrum Disorder (自閉症的致病機轉)
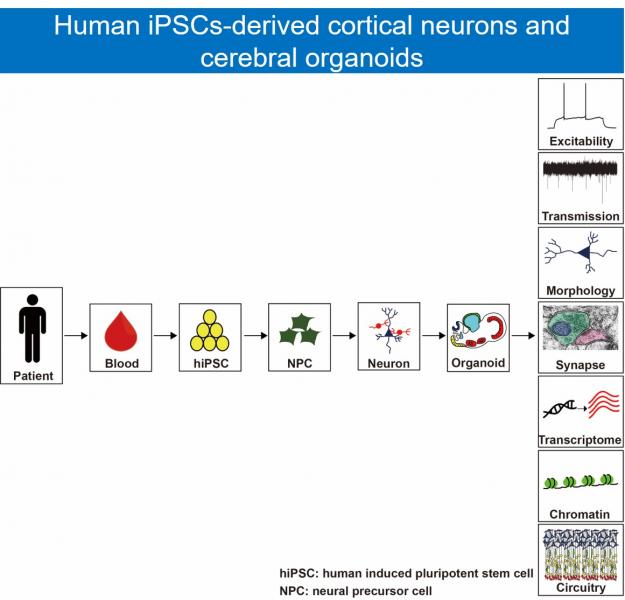
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by dysfunctions in social interactions, communication difficulties, and repetitive patterns of behavior. ASD affects one out of every 59 children. The exact causal mechanism for ASD is still unknown, therefore, there is no effective treatment for ASD. We previously identified mono-to-biallelic switch of autism susceptibility genes from persons with ASD1. To further study the underlying causal mechanisms of ASD, we use human induced pluripotent stem cells (hiPSCs)-derived cortical neurons and cerebral organoids as research tools. Our research goal is to uncover the pathogenesis of ASD.
自閉症為一個盛行率極高的兒童神經發育疾病,其致病機轉仍尚未清楚,目前也無有效的治療藥物。我們利用病人血球細胞所分化出來的活體神經細胞及大腦類器官,來研究自閉症的致病機轉。
Reference:
自閉症為一個盛行率極高的兒童神經發育疾病,其致病機轉仍尚未清楚,目前也無有效的治療藥物。我們利用病人血球細胞所分化出來的活體神經細胞及大腦類器官,來研究自閉症的致病機轉。
Reference:
1. Lin CY, Chang KW, Lin CY, Wu JY, Coon H, Huang PH, Ho HN, Akbarian S, Gau SS, Huang HS. (2018) Allele-specific expression in a family quartet with autism reveals mono-to-biallelic switch and novel transcriptional processes of autism susceptibility genes. Scientific Reports 8:4277.
Mechanisms and Functions of Brain Imprintome (基因組印記在大腦形成的機轉及功能)
Epigenetic machinery can change gene expression without altering the genetic sequence. Genomic imprinting, which is particularly important in the nervous system (NS), is an epigenetic process through which monoallelic gene expression occurs in a parent-of-origin-specific manner. Dysregulation of imprinted genes causes various neurological and psychiatric disorders. Despite its importance in brain disorders, the role of genomic imprinting in the NS remains unclear. Accordingly, there is a need to characterize imprinted genes in the NS and to understand their underlying biology.
To address this critical question, we comprehensively profiled and validated imprinted genes, long non-coding RNAs and microRNAs, in the mouse visual system1 and human prefrontal cortex2. In addition to those findings, we identified several novel genomic imprinting phenomena in the specific brain regions mentioned above in mouse and human. We also built up a multi-stage platform to study gene imprinting in a single-cell type level3. To further investigate the role of genomic imprinting in the brain, we currently focus on studying the mechanisms and functions of brain imprintome4-8. Our research would provide new insights into the regulation of genomic imprinting in the context of psychiatric and neurological diseases, due to the association of these diseases with dysfunctional genomic imprinting.
父方及母方來源的染色體,在功能上是不相同的,其差異性會造成基因組印記。基因組印記盛行於大腦,其在大腦的功能尚未清楚,但其缺失會導致許多大腦疾病。目前我們聚焦在研究基因組印記在大腦中,形成的機轉及其生理功能。
Reference:
To address this critical question, we comprehensively profiled and validated imprinted genes, long non-coding RNAs and microRNAs, in the mouse visual system1 and human prefrontal cortex2. In addition to those findings, we identified several novel genomic imprinting phenomena in the specific brain regions mentioned above in mouse and human. We also built up a multi-stage platform to study gene imprinting in a single-cell type level3. To further investigate the role of genomic imprinting in the brain, we currently focus on studying the mechanisms and functions of brain imprintome4-8. Our research would provide new insights into the regulation of genomic imprinting in the context of psychiatric and neurological diseases, due to the association of these diseases with dysfunctional genomic imprinting.
父方及母方來源的染色體,在功能上是不相同的,其差異性會造成基因組印記。基因組印記盛行於大腦,其在大腦的功能尚未清楚,但其缺失會導致許多大腦疾病。目前我們聚焦在研究基因組印記在大腦中,形成的機轉及其生理功能。
Reference:
1. Hsu CL, Chou CH, Huang SC, Lin CY, Lin MY, Tung CC, Lin CY, Lai IP, Zou YF, Youngson NA, Lin SP, Yang CH, Chen SK, Gau SS, Huang HS. (2018) Analysis of experience-regulated transcriptome and imprintome during critical periods of mouse visual system development reveals spatiotemporal dynamics. Human Molecular Genetics 27(6):1039-1054.
2. Lin CY, Chang KW, Lin CY, Wu JY, Coon H, Huang PH, Ho HN, Akbarian S, Gau SS, Huang HS. (2018) Allele-specific expression in a family quartet with autism reveals mono-to-biallelic switch and novel transcriptional processes of autism susceptibility genes. Scientific Reports 8:4277.
3. Lin CY, Huang SC, Tung CC, Chou CH, Gau SS, Huang HS. (2016) Analysis of Genome-Wide Monoallelic Expression Patterns in Three Major Cell Types of Mouse Visual Cortex Using Laser Capture Microdissection. PLOS ONE 11(9):e0163663.
4. Chou MY, Appan D, Chang KW, Chou CH, Lin CY, Gau SS, Huang HS. (2022) Mouse hybrid genome mediates diverse brain phenotypes with the specificity of reciprocal crosses. FASEB J 36(3):e22232.
5. Chou MY, Hu MC, Chen PY, Hsu CL, Lin TY, Tan MJ, Lee CY, Kuo MF, Huang PH, Wu VC, Yang SH, Fan PC, Huang HY, Akbarian S, Loo TH, Stewart CL, Huang HP, Gau SS, Huang HS. (2022) RTL1/PEG11 imprinted in human and mouse brain mediates anxiety-like and social behaviors and regulates neuronal excitability in the locus coeruleus. Human Molecular Genetics 31(18):3161-3180.
6. Chou MY, Cao X, Hou KC, Tsai MH, Lee CY, Kuo MF, Wu VC, Huang HY, Akbarian S, Chang SK, Hu CY, Lin SW, Huang HS. (2023) Mir125b-2 imprinted in human but not mouse brain regulates hippocampal function and circuit in mice. Communications Biology 6:267.
7. Hou KC, Tsai MH, Akbarian S, Huang HS. (2023) Mir125b-1 is not imprinted in human brain and shows developmental expression changes in mouse brain. Neuroscience 529:99-106.
8. Lin CW, Cheng YC, Yang CH, Huang HS. (2023) Light activates Ube3a, an Angelman syndrome-associated gene, by mediating the chromatin structures during postnatal development of mouse retina. Journal of Neurochemistry Dec;167(6):766-777.
4. Chou MY, Appan D, Chang KW, Chou CH, Lin CY, Gau SS, Huang HS. (2022) Mouse hybrid genome mediates diverse brain phenotypes with the specificity of reciprocal crosses. FASEB J 36(3):e22232.
5. Chou MY, Hu MC, Chen PY, Hsu CL, Lin TY, Tan MJ, Lee CY, Kuo MF, Huang PH, Wu VC, Yang SH, Fan PC, Huang HY, Akbarian S, Loo TH, Stewart CL, Huang HP, Gau SS, Huang HS. (2022) RTL1/PEG11 imprinted in human and mouse brain mediates anxiety-like and social behaviors and regulates neuronal excitability in the locus coeruleus. Human Molecular Genetics 31(18):3161-3180.
6. Chou MY, Cao X, Hou KC, Tsai MH, Lee CY, Kuo MF, Wu VC, Huang HY, Akbarian S, Chang SK, Hu CY, Lin SW, Huang HS. (2023) Mir125b-2 imprinted in human but not mouse brain regulates hippocampal function and circuit in mice. Communications Biology 6:267.
7. Hou KC, Tsai MH, Akbarian S, Huang HS. (2023) Mir125b-1 is not imprinted in human brain and shows developmental expression changes in mouse brain. Neuroscience 529:99-106.
8. Lin CW, Cheng YC, Yang CH, Huang HS. (2023) Light activates Ube3a, an Angelman syndrome-associated gene, by mediating the chromatin structures during postnatal development of mouse retina. Journal of Neurochemistry Dec;167(6):766-777.
.jpg)
Alternative RNA Splicing in the Brain Function (選擇性RNA剪接在大腦的功能)
Alternative RNA splicing is active during neurodevelopment, however, its biological functions in the brain are under investigation. RBFOX3 (RNA binding protein, fox-1 homolog (C. elegans) 3) is a neuron-specific alternative splicing regulator. Disruptions of RBFOX3 have been identified in persons with epilepsy, cognitive impairments, attention-deficit hyperactivity disorder, and sleep problems. Our previous work showed a causal link between the disruption of Rbfox3 and epilepsy and cognitive impairments1. Moreover, we demonstrated that RBFOX3 is required for hippocampal circuit balance and function1, in addition to adult hippocampal neurogenesis and synaptogenesis2. We also showed that RBFOX3 is dispensable for visual function, despite its specific expression in the ganglion cells of the retina3. We currently focus on investigating the mechanisms for how dysfunctional RBFOX3 contributes to epilepsy4, cognitive impairments, attention-deficit hyperactivity disorder, and sleep problems. Our research goal is to identify novel and unexpected mechanisms and physiological functions of alternative RNA splicing in the brain.
選擇性RNA剪接,活躍於大腦中,然而其在大腦中的生理功能,仍尚未清楚。調控大腦選擇性RNA剪接的其中關鍵分子之一為RBFOX3,其缺失會導致癲癇、認知障礙、注意力不足過動症及睡眠障礙。我們目前專注於研究RBFOX3如何導致其相關疾病的機轉。
Reference:
1. Wang HY, Hsieh PF, Huang DF, Chin PS, Chou CH, Tung CC, Chen SY, Lee LJ, Gau SS, Huang HS. (2015) RBFOX3/NeuN is required for hippocampal circuit balance and function. Scientific Reports 5, 17383.
2. Lin YS, Wang HY, Huang DF, Hsieh PF, Lin MY, Chou CH, Wu IJ, Huang GJ, Gau SS, Huang HS. (2016) Neuronal Splicing Regulator RBFOX3 (NeuN) Regulates Adult Hippocampal Neurogenesis and Synaptogenesis. PLOS ONE 11(10): e0164164.
3. Lin YS, Kuo KT, Chen SK, Huang HS. (2018) RBFOX3/NeuN is dispensable for visual function. PLOS ONE 13(2): e0192355.
4. Huang DF, Lee CY, Chou MY, Yang TY, Cao X, Hsiao YH, Wu RN, Lien CC, Huang YS, Huang HP, Gau SS, Huang HS. (2022) Neuronal splicing regulator RBFOX3 mediates seizures via regulating Vamp1 expression preferentially in NPY-expressing GABAergic neurons. Proc Natl Acad Sci U S A. 119(33):e2203632119.
選擇性RNA剪接,活躍於大腦中,然而其在大腦中的生理功能,仍尚未清楚。調控大腦選擇性RNA剪接的其中關鍵分子之一為RBFOX3,其缺失會導致癲癇、認知障礙、注意力不足過動症及睡眠障礙。我們目前專注於研究RBFOX3如何導致其相關疾病的機轉。
Reference:
1. Wang HY, Hsieh PF, Huang DF, Chin PS, Chou CH, Tung CC, Chen SY, Lee LJ, Gau SS, Huang HS. (2015) RBFOX3/NeuN is required for hippocampal circuit balance and function. Scientific Reports 5, 17383.
2. Lin YS, Wang HY, Huang DF, Hsieh PF, Lin MY, Chou CH, Wu IJ, Huang GJ, Gau SS, Huang HS. (2016) Neuronal Splicing Regulator RBFOX3 (NeuN) Regulates Adult Hippocampal Neurogenesis and Synaptogenesis. PLOS ONE 11(10): e0164164.
3. Lin YS, Kuo KT, Chen SK, Huang HS. (2018) RBFOX3/NeuN is dispensable for visual function. PLOS ONE 13(2): e0192355.
4. Huang DF, Lee CY, Chou MY, Yang TY, Cao X, Hsiao YH, Wu RN, Lien CC, Huang YS, Huang HP, Gau SS, Huang HS. (2022) Neuronal splicing regulator RBFOX3 mediates seizures via regulating Vamp1 expression preferentially in NPY-expressing GABAergic neurons. Proc Natl Acad Sci U S A. 119(33):e2203632119.
.jpg)
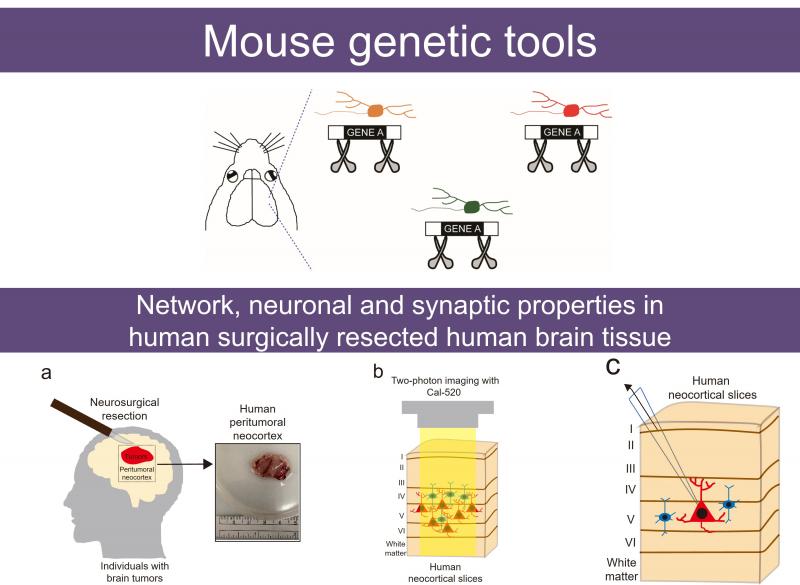
Developing new research tools (開發新的研究工具)
We are currently developing new tools to enable more powerful approaches to our research questions. Specifically, we are focusing on mice which can delete target genes in cell-subtype-1, isoform-, and parent-of-origin-specific manners. Moreover, we established a robust platform to assess population-wide neuronal activity and single-cell neuronal and synaptic properties in surgically resected human brain tissue2.
我們致力於開發新的研究工具,以讓我們可以更精準地回答我們所關心的研究問題。目前,我們正專注於開發能夠剔除具細胞亞型、異構體及父母染色體來源專一性基因的工程小鼠。此外,我们已經建立了一套稳健的平台,可用于分析经外科切除的人類腦組織中,整體神经元活动,以及單细胞層级的神经元與突觸特性。
Reference:
1.Huang DF, Lin CW, Yang TY, Lien CC, Yang CH, Huang HS. (2023) An intersectional genetic approach for simultaneous cell-type-specific labeling and gene knockout in the mouse. Development Feb 15;150(4):dev201198.
2.Huang DF, Chou MY, Lee YH, Chen YT, Sun CK, Wang KC, Huang HS. (2025) Subarachnoid hemorrhage mediates human neocortical network, membrane potential, and action potential bursting via glutamate receptors. Communications Biology Nov 27;8(1):1710.
我們致力於開發新的研究工具,以讓我們可以更精準地回答我們所關心的研究問題。目前,我們正專注於開發能夠剔除具細胞亞型、異構體及父母染色體來源專一性基因的工程小鼠。此外,我们已經建立了一套稳健的平台,可用于分析经外科切除的人類腦組織中,整體神经元活动,以及單细胞層级的神经元與突觸特性。
Reference:
1.Huang DF, Lin CW, Yang TY, Lien CC, Yang CH, Huang HS. (2023) An intersectional genetic approach for simultaneous cell-type-specific labeling and gene knockout in the mouse. Development Feb 15;150(4):dev201198.
2.Huang DF, Chou MY, Lee YH, Chen YT, Sun CK, Wang KC, Huang HS. (2025) Subarachnoid hemorrhage mediates human neocortical network, membrane potential, and action potential bursting via glutamate receptors. Communications Biology Nov 27;8(1):1710.